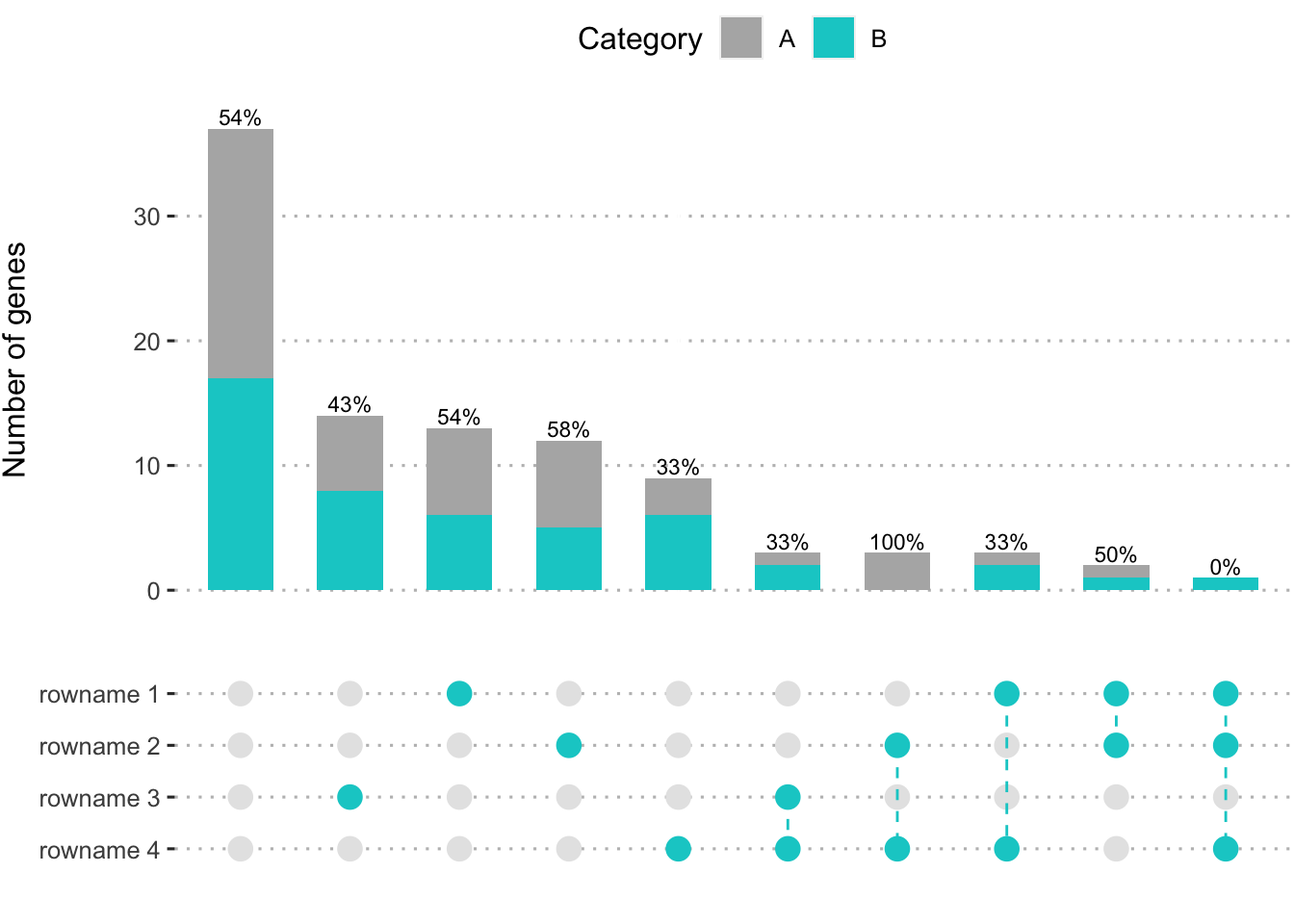
Rebuild UpSet plot color
UpSet plot is a ready-to-use package for visualizing categorical overlaps. But it can be tricky to modify its barplot color and add the bar subclass to allow the additional comparison.
Here, I made the UpSet style plot, which can change color and number of intersections, and keep the top interactions for simplicity.
library(ggplot2)
library(cowplot)
plot_freq_intersect <- function(dat, .by = "Group", .levels = NA, .split = "Category", .color = 1:10, top_n = 10) {
# Args:
# dat: gene_id (unique)
# .by: row feature
# .levels: dot plot row names
# .split: bar split feature
if (is.na(.levels[1])) {
.levels <- seq_len(as.character(nchar(dat[1, .by])))
}
# limit less frequent groups
top_groups <- names(tail(sort(table(dat[, .by])), top_n))
dat <- dat[dat[[.by]] %in% top_groups, ]
dat$Group <- factor(dat[, .by], levels = names(sort(table(dat[, .by]), decreasing = T)))
dat$Type <- dat[, .split]
# barplot
dat_g1 <- dplyr::count(dat, Group, Type, sort = TRUE)
frac_tbl <- table(dat[, "Group"], dat[, "Type"])
frac_tbl <- frac_tbl / rowSums(frac_tbl)
dat_g1_text <- data.frame(Group = rownames(frac_tbl),
Fraction = paste0(round(frac_tbl[, 1], 2) * 100, "%"),
Type = colnames(frac_tbl)[1])
Group_n <- dplyr::count(dat, Group, sort = TRUE)
dat_g1_text$n <- Group_n[match(dat_g1_text$Group, Group_n$Group), "n"]
g1 <- ggplot(dat_g1, aes(x = Group, y = n, fill = Type)) +
geom_bar(stat = "identity", width = 0.6) +
geom_text(data = dat_g1_text,
aes(x = Group, y = n, label = Fraction),
vjust = -0.25, hjust = 0.5,
size = 3) +
xlab("") + ylab("Number of genes") +
ggpubr::theme_pubclean() +
scale_fill_manual(name = .split, values = c("grey70", .color)) +
theme(axis.text.x = element_blank(),
axis.ticks.x = element_blank(),
plot.margin = margin(c(0,1,1,1)))
# dot plot
dat_tile <- NULL
for (m in seq_along(levels(dat$Group))) {
tmp <- cbind(x = m, y = .levels,
color = ifelse(strsplit(levels(dat$Group)[m], "")[[1]] == 1, 1, 0))
dat_tile <- rbind(dat_tile, tmp)
}
dat_tile <- as.data.frame(dat_tile)
dat_tile$x <- factor(dat_tile$x, levels = order(as.numeric(dat_tile$x)))
dat_tile$y <- factor(dat_tile$y, levels = rev(.levels))
g2 <- ggplot(dat_tile, aes(x = x, y = y, color = factor(color), group = x)) +
geom_point(size = 4) +
scale_color_manual(values = c("grey90", .color)) +
xlab("") + ylab("") +
ggpubr::theme_pubclean() +
theme(panel.grid = element_blank(),
legend.position = "none",
axis.text.x = element_blank(),
axis.ticks.x = element_blank(),
plot.margin = margin(c(1,1,0,1)))
# add lines
dat_seg <- NULL
for (i in unique(dat_tile$x)) {
tmp <- dat_tile[dat_tile$x == i, ]
tmp <- tmp[tmp$color == 1, ]
if (nrow(tmp) > 1) {
dat_seg <- rbind(dat_seg,
data.frame(x = tmp$x[-nrow(tmp)], y = tmp$y[-nrow(tmp)],
xend = tmp$x[-1], yend = tmp$y[-1]))
}
}
if (!is.null(dat_seg)) {
levels(dat_seg$x) <- levels(dat_seg$xend) <- levels(dat_tile$x)
levels(dat_seg$y) <- levels(dat_seg$yend) <- levels(dat_tile$y)
dat_seg$color <- 1
g2 <- g2 + geom_segment(data = dat_seg, aes(x = x, y = y, xend = xend, yend = yend), lty = 2)
}
cowplot::plot_grid(g1, g2, ncol = 1, align = "v", rel_heights = c(1, 0.4))
}Below is a toy data set for testing.
gene_set_1 <- sample(100, 20, replace = FALSE)
gene_set_2 <- sample(100, 20, replace = FALSE)
gene_set_3 <- sample(100, 20, replace = FALSE)
gene_set_4 <- sample(100, 20, replace = FALSE)
dat_test <- data.frame(gene_id = 1:100,
Category = rep(c("A", "B"), 50))
# convert to group of intersection
dat_test$Group <- paste0(as.numeric(dat_test$gene_id %in% gene_set_1),
paste0(as.numeric(dat_test$gene_id %in% gene_set_2),
paste0(as.numeric(dat_test$gene_id %in% gene_set_3),
as.numeric(dat_test$gene_id %in% gene_set_4))))
plot_freq_intersect(dat_test, .by = "Group",
.levels = c("rowname 1", "rowname 2", "rowname 3", "rowname 4"),
.split = "Category", .color = "cyan3", top_n = 10)