Does steady-state appear in a growing cell?
2022 Oct 24
RNA turnover
Single cell
Thought
Cellular RNA is balanced by RNA synthesis and degradation, as a water container with an input and an outlet (or many outlets if total RNA proportionally decays).
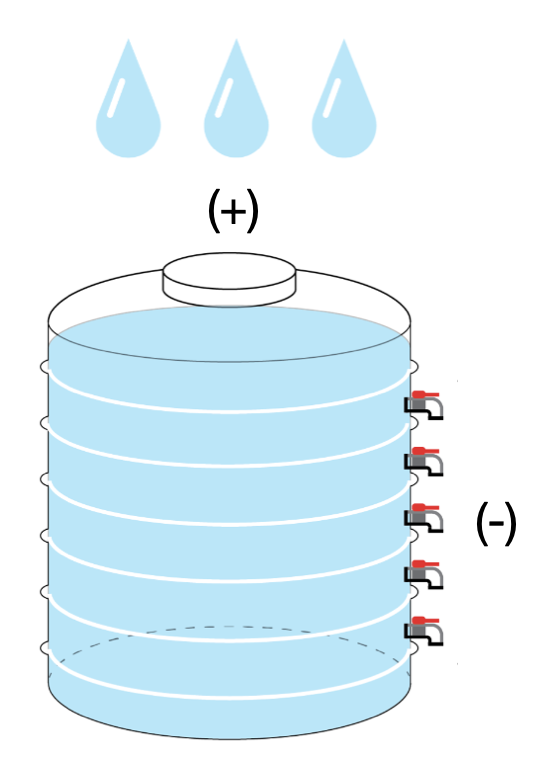
Total RNA abundance from synthsis and decay balance.
In terms of RNA metabolic rate, RNA half-life describes how long half of the current RNA can be replaced or disposed. However, calculating this parameter often requires the “steady-state hypothesis”, which assumes that RNA synthesis and RNA decay are the same.
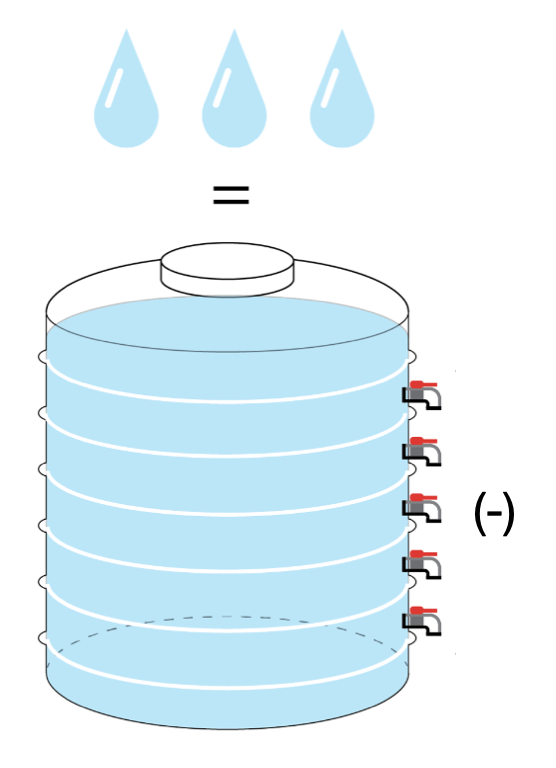
Total RNA turnover in the “steady-state”.
This hypothesis simplifies the RNA half-life calculation, originating from bulk RNA-seq analysis. But, at a single-cell level, “steady-state” contradicts with cell growth. If RNA synthesis must equal its decay, total RNA would be halved by every cell division, resulting an increasingly smaller RNA abundance.
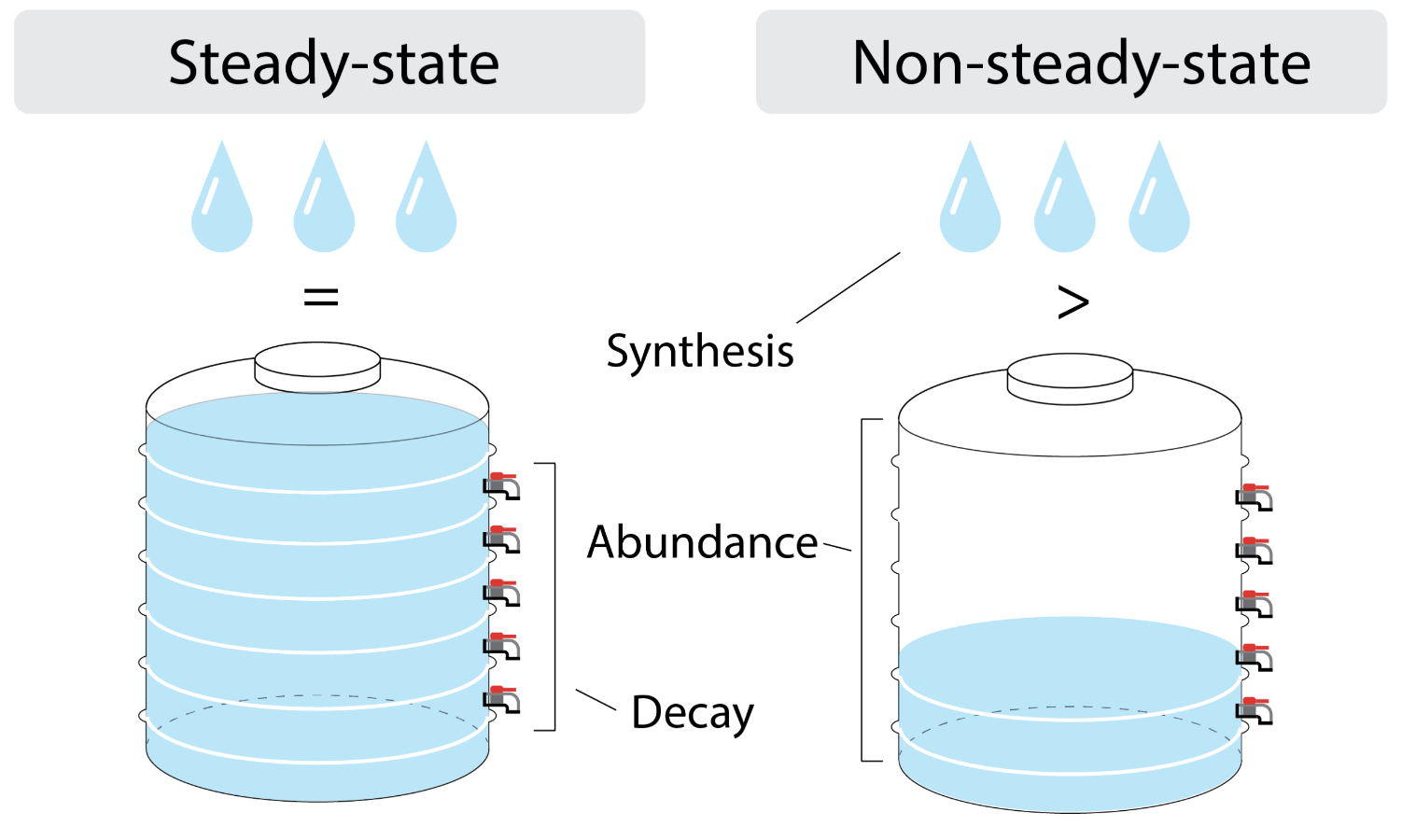
Total RNA turnover in the “non-steady-state”.
If assuming cells are in a “non-steady-state”, everything will make sense. First, the unsaturated cellular RNA allows total RNA to accumulate in the subsequent cell cycle. And, gene expression regulation can easily occur with transcriptional changes, when RNA degradation plays a minor role.
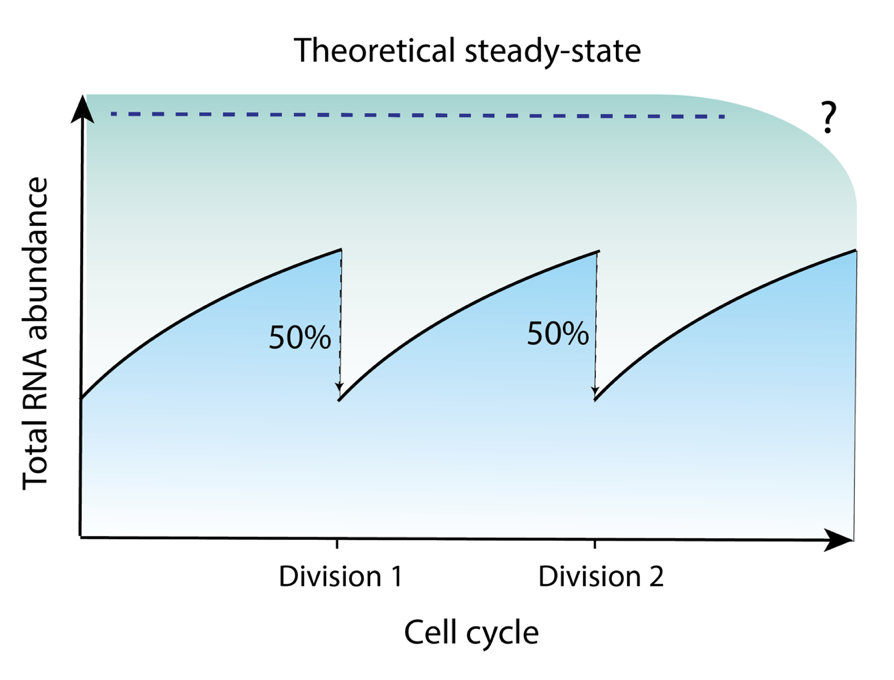
Total RNA abundance changes across cell cycles.
Currently, RNA metabolic labeling sequencing measures absolute RNA synthesis rate, and TT-seq shows shorter RNA turnover half-lives (a median ~1 hour, mESCs in serum medium) than RNA absolute degradation half-lives (~4 hours, in SLAM-seq and TimeLapse-seq, serum medium). The faster turnover suggests RNA synthesis can renew the current total RNA easily before the pre-existing RNA shattering via degradation. So for cells in rapid growth (e.g. mESCs), the “non-steady-state” is more likely to appear.
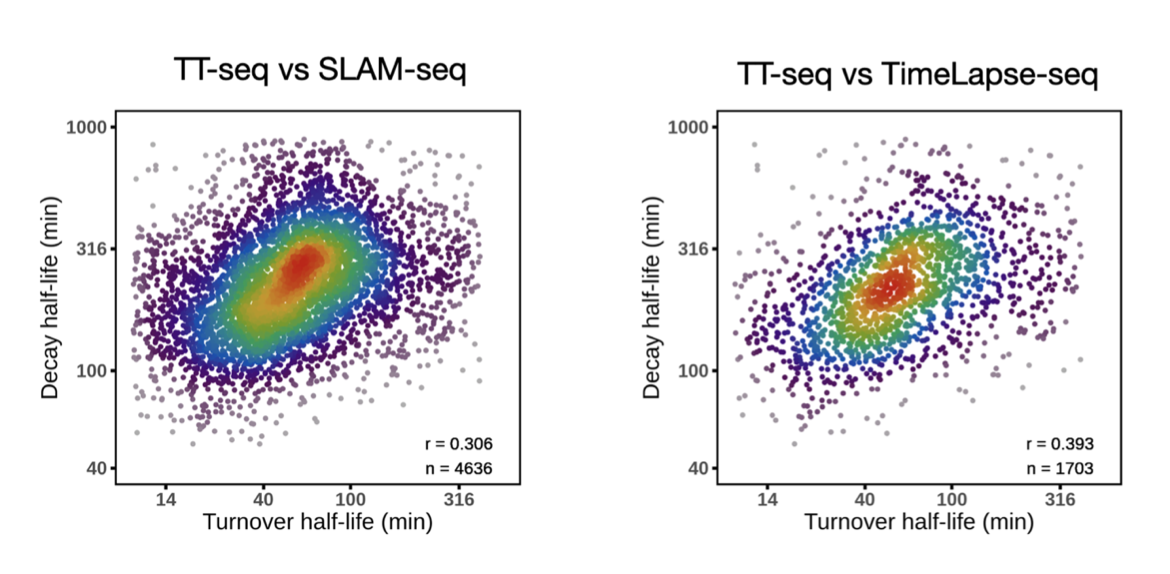
RNA turnover half-life (TT-seq) vs RNA decay half-life (SLAM-seq, TimeLapse-seq)
In addition, a single-cell metabolic labeling (5-EU) study has found that the “non-steady-state” showed better model fitting (Battich et al. 2020, Science). This study highlights the need to adjust the RNA “steady-state” hypothesis to a new dynamic turnover strategy especially for single-cell studies.
If you found the idea above useful for your study, please cite the related paper: “Distinct transcription kinetics of pluripotent cell states”.